We use a combination of quantitative experiments and mathematical modeling to study signaling dynamics and spatial cellular organization in early embryonic development. Research focuses on two major related areas:
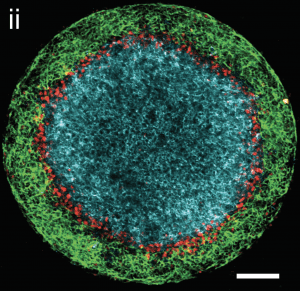
A system which recapitulates the dorsal-ventral patterning of human ectoderm. Shown are neural (blue), neural crest (red), and surface ectodermal (green) fates. (Britton et al. bioRxiv 2018).
1. Self-organized developmental patterning. Developmental biologists have long marveled at the ability of the embryo, a single cell without clear structure, to self-organize into an entire organism. Over the past thirty years, revolutions in biochemistry and molecular biology have allowed for the identification of genes involved in self-organization and patterning, and shown that many of these encode secreted ligands and inhibitors, known as morphogen signals, which control cell fate. Despite this progress, a mechanistic understanding of how pattern emerges has remained largely elusive. We use human embryonic stem cells to create systems in which cells self-organize into patterns that are reminiscent of early human development. These systems allow us to study how these patterns form using perturbative experiments and imaging approaches. We currently work with two systems: one with recapitulates gastrulation, the earliest decision the cells of the embryo proper make, and another which models the patterning of the human ectoderm into four distinct territories along its dorsal-ventral axis. These systems allow us to address fundamental questions and to study human development directly.
Key Publications: Warmflash et al. Nature Methods 2014, Nemashkalo et al Development 2017, Chhabra et al. bioRxiv 2018, Britton et al. bioRxiv 2019.
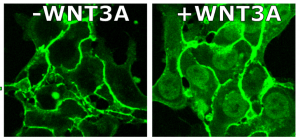
Reporter line in human embryonic stem cells with GFP integrated at the endogenous b-catenin locus, allows us to monitor Wnt signaling in real time (Massey et al PNAS 2019)
2. Dynamic processing by morphogen signaling pathways. During development, cells migrate and expression patterns of signaling ligands change more rapidly than the timescale for cells to make fate decision. This means that classic models which involve cells reading a fixed gradient of ligand concentration must be oversimplified and cells must dynamically interpret the signals that instruct them to adopt cell fates. While the biochemistry of many key signaling pathways is understood in great detail, knowledge of how morphogen signaling pathways respond to dynamically varying ligands remains limited. Work in the laboratory uses live-cell reporters in human embryonic stem cells to probe the dynamics of signaling pathways with a focus on the TGF-beta superfamily and Wnt pathways. This approach allows us to understand how the dynamic properties of each pathway are utilized during develpment in order to drive cell fate decisions.
Key publications: Sorre et al Dev Cell 2014, Heemskerk et a eLife 2019, Massey et al PNAS 2019.